I was revising my statistics and data analytics notes from my dog eared handwritten notebooks and thought it would be a good idea to transfer the notes online. What better place than the blog.
Here is a quick and simple example of the KMeans Clustering algorithm. And to demonstrate the algo, I am using the infamous IRIS dataset. I do apologise if you are bored at looking at this dataset over and over again, but its probably the most simplest and easily understandable dataset for beginners.
The data set has 150 rows, with 4 columns/features describing the Sepal Length, Sepal Width, Petal Length, Petal Width of three different species of the Iris flower. The data set has a 5th column that identifies what the species is. We will look at the structure of the dataset further down.
To understand any data analytics concept, we should start with a question that we want the data to answer for us. The question I have is, “Is there a pattern in the data, whereby we can group the three species of Iris, so that if we see a new sample, we could identify what species it belongs to?”
We want a way to group the samples automatically, without us ‘training’ the algorithm. i.e., we are looking for an Unsupervised Learning algorithm.
KMeans Clustering is one such Unsupervised Learning algo, which, by looking at the data, groups the samples into ‘clusters’ based on how far each sample is from the group’s centre.
A bit more info on KMeans is here.
Right, let’s dive right in and see how we can implement KMeans clustering in Python.
You would need the following packages installed:
- sklearn (https://pypi.python.org/pypi/scikit-learn)
- pandas (https://pypi.python.org/pypi/pandas)
- numpy (http://www.numpy.org/)
- matplotlib (http://matplotlib.org/)
Conveniently, sklearn package comes with a bunch of useful datasets. One of them is Iris data.
Import the packages
from sklearn import datasets from sklearn.cluster import KMeans import pandas as pd import numpy as np import matplotlib.pyplot as plt
Load the iris data and take a quick look at the structure of the data. The sepal and petal lengths and widths are in an array called iris.data. The species classifications for each of the 150 samples is in another array called iris.target.
iris = datasets.load_iris() print iris.data print iris.target
Output: array([[ 5.1, 3.5, 1.4, 0.2],
[ 4.9, 3. , 1.4, 0.2],
[ 4.7, 3.2, 1.3, 0.2],
[ 4.6, 3.1, 1.5, 0.2],
[ 5. , 3.6, 1.4, 0.2],
[ 5.4, 3.9, 1.7, 0.4],
....
Output: array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2])
Let’s convert our arrays to a pandas DataFrame for ease of use. I am setting the column names explicitly.
x = pd.DataFrame(iris.data, columns=['Sepal Length', 'Sepal Width', 'Petal Length', 'Petal Width']) y = pd.DataFrame(iris.target, columns=['Target'])
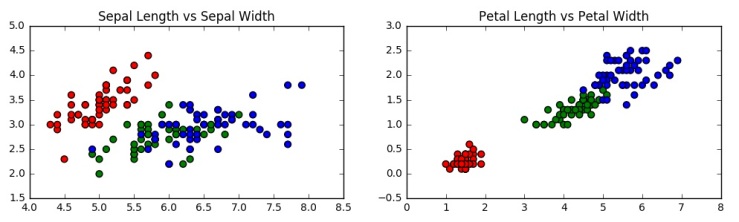
Now, lets quickly visualise the data in a scatter plot to see if there is any pattern visible.
# Start with a plot figure of size 12 units wide & 3 units tall
plt.figure(figsize=(12,3))
# Create an array of three colours, one for each species.
colors = np.array(['red', 'green', 'blue'])
#Draw a Scatter plot for Sepal Length vs Sepal Width
#nrows=1, ncols=2, plot_number=1
# http://matplotlib.org/api/pyplot_api.html#matplotlib.pyplot.subplot
plt.subplot(1, 2, 1)
# http://matplotlib.org/api/pyplot_api.html#matplotlib.pyplot.scatter
plt.scatter(x['Sepal Length'], x['Sepal Width'], c=colors[y['Target']], s=40)
plt.title('Sepal Length vs Sepal Width')
plt.subplot(1,2,2)
plt.scatter(x['Petal Length'], x['Petal Width'], c= colors[y.Targets], s=40)
plt.title('Petal Length vs Petal Width')
We can clearly see the grouping in the plots with the red dots, which correspond to species Setosa. The green and blue dots are not so clearly separable.
Now let’s use the KMeans algorithm to see if it can create the clusters automatically.
# create a model object with 3 clusters # http://scikit-learn.org/stable/modules/generated/sklearn.cluster.KMeans.html#sklearn.cluster.KMeans # http://scikit-learn.org/stable/modules/clustering.html#k-means model = KMeans(n_clusters=3) model.fit(x)
model.fit() function runs the algo on the data and creates the clusters. Each sample in the dataset is then assigned a cluster id (0, 1, 2, etc).
model.labels_ holds the array of the cluster ids, so let’s take a look at it.
print model.labels_
#Start with a plot figure of size 12 units wide & 3 units tall
plt.figure(figsize=(12,3))
# Create an array of three colours, one for each species.
colors = np.array(['red', 'green', 'blue'])
# The fudge to reorder the cluster ids.
predictedY = np.choose(model.labels_, [1, 0, 2]).astype(np.int64)
# Plot the classifications that we saw earlier between Petal Length and Petal Width
plt.subplot(1, 2, 1)
plt.scatter(x['Petal Length'], x['Petal Width'], c=colormap[y['Target']], s=40)
plt.title('Before classification')
# Plot the classifications according to the model
plt.subplot(1, 2, 2)
plt.scatter(x['Petal Length'], x['Petal Width'], c=colormap[predictedY], s=40)
plt.title("Model's classification")
We can see that all the red dots are grouped/clustered 100% accurately and the green and black dots are fairly well grouped too.
So there we have it. We have used the KMeans clustering algorithm to automatically group the 150 samples of Iris flowers into three clusters.
Edit: I learn a lot from the fellow bloggers out there (thank you all) and I learnt this clustering example from John Stamford’s blog post. Thanks again.



Thx for the detailed post !! It’s well written . If I may ask u a quick question on this please , I have my own table of master data of some skus and they have attributes like product group , size , color , description.
Can I just convert this to a pandas data frame and fit into the model ?
And the run a model.fit()
Would I see anything meaningful and would be the right way to do it ?
LikeLike
useful information and than python coding
LikeLike